A short explanation is given for each motif annotated by NASSAM and base interaction clusters annotated by COGNAC
A base-triple is a widespread tertiary interaction that consists of three bases interacting via hydrogen bonds in a planar orientation. For triples in the NASSAM database, each nucleotide base is interacting with other bases by at least two hydrogen bonds. Majority of the base-triple patterns were built from the Nucleic Acid Interaction Library (NAIL).
References 1) Walberer BJ, Cheng AC, Frankel AD. 2003. Structural diversity and isomorphism of hydrogen-bonded base interactions in nucleic acids. J Mol Biol 327: 767-780. 2) Firdaus-Raih M, Harrison AM, Willett P, and Artymiuk PJ. (2011) Novel base triples in RNA structures revealed by graph theoretical searching methods. BMC Bioinformatics, 12(S13): S2. A/G-MinorsThe A-minor motif is formed when an adenosine interacts with the minor-groove of a receptor base pair, which is usually a Watson-Crick base pair. The A-minor motif can be divided into 4 types based on the position of the inserted adenosine relative to the receptor base pair. G-minor is a variant of the A-minor motif in which guanosine docks to the minor groove of a receptor base-pair instead of an adenosine. At present InterRNA contains annotation for Type I A-minors and Type II A/G-minors only.
Reference 1) Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. PNAS 98: 4899-4903. Ribose-ZippersThe ribose-zipper interaction is characterized by consecutive hydrogen-bonding interactions between ribose 2'-hydroxyls from different regions of an RNA chain or between RNA chains.
Reference 1) Tamura M, Holbrook SR. 2002. Sequence and structural conservation in RNA ribose zippers. J Mol Biol 320: 455-474. TetraloopsA tetraloop is a four-nucleotide hairpin loop in RNA secondary structure that caps many helices. They can generally be divided into three sequence motifs: the 'UNCG', the 'GNRA' and the 'CUYG' tetraloops. In these sequences, R stands for a purine base (R= A or G), Y stands for a pyrimidine base (Y = U or C) and N indicates the position that can be any base. (N = A, U, G, C).
NOTE: All tetraloop hits are filtered on the basis of having 4 consecutive nucleotides. This is done to prevent the occurrences of false positives and to maintain the quality of annotations in InterRNA. One caveat of this approach is that it sometimes misses tetraloops that have inserted or bulged out nucleotides (longer than 4 nucleotides). Work is in progress to annotate these interactions and provide them through InterRNA.
Reference 1) Correll CC, Swinger K. 2005. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 resolution. RNA 9: 355-363 2) Jucker FM, Pardi A. 1995. Solution structure of the CUUG hairpin loop: a novel RNA tetraloop motif. Biochemistry 34: 14416-14427 3) Cheong C, Varani G, Tinoco Jr I. 1990. Solution structure of an unusually stable RNA hairpin 5'GGAC(UUCG)GUCC. Nature 346: 680-682 T-LoopsThe T-loop, originally characterized in tRNA is a five-nucleotide hairpin-loop motif composed of a U-turn flanked by a non-canonical base pair.
NOTE: All T-loop hits are filtered on the basis of having 5 consecutive nucleotides. Similar to tetraloops, T-loops are also capable of accommodating inserted or bulged out nucleotides making the length of some of these interactions to be more than 5 nucleotides. Work is in progress to annotate these interactions and provide them through InterRNA.
Reference 1) Krasilnikov AS, Mondragon A. 2003. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA 9: 640-643. Base Interaction ClustersThe base interaction clusters annotated by COGNAC can be represented by tree graphs where each base has at least one hydrogen bond connection as part of a network in a cluster of three to six bases (see below).
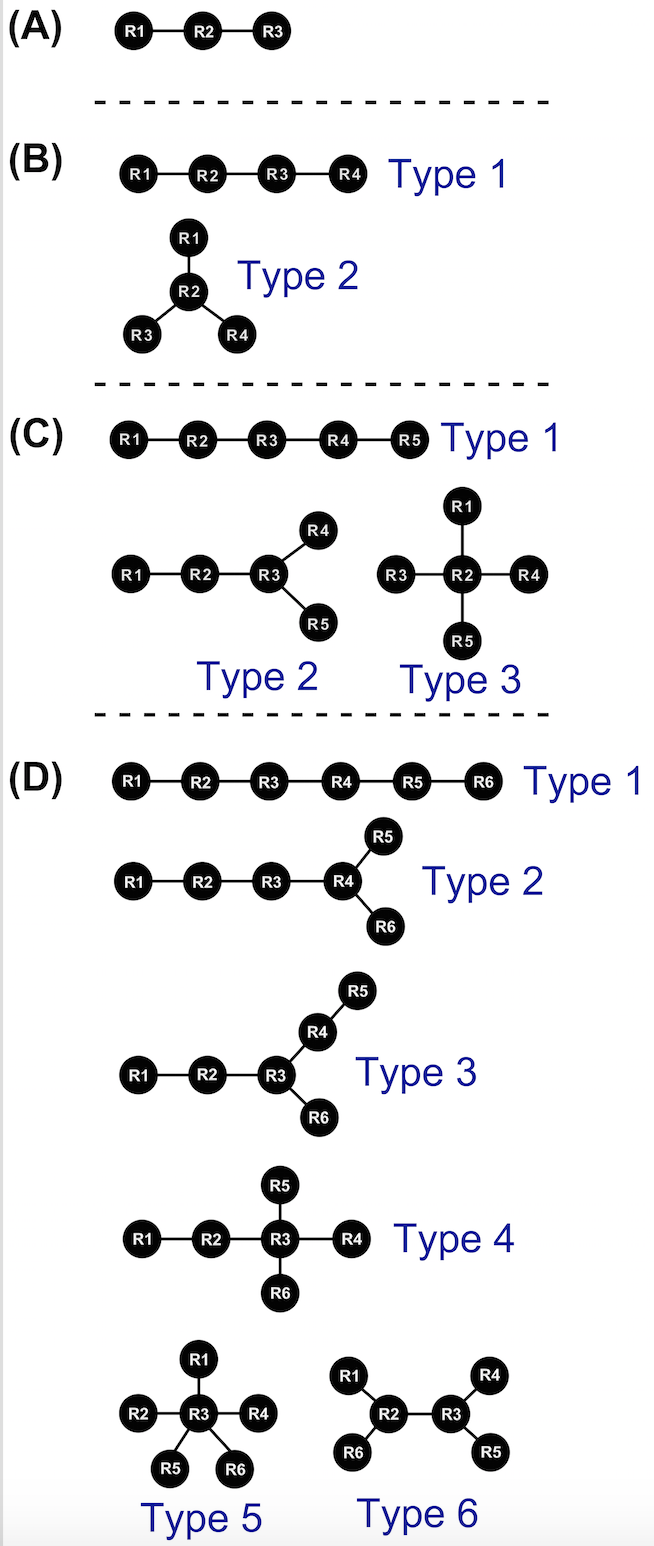 Figure above presents the tree graphs used by COGNAC to represent hydrogen bond connected base interactions where R denotes a base residue position (node) and the edges of the graphs are the lines connecting the nodes for: (A) triples (B) quadruples (C) quintuples (D) sextuples.
Reference
1) Mohd Firdaus-Raih, Hazrina Yusof Hamdani, Nurul Nadzirin, Effirul Ikhwan Ramlan, Peter Willett and Peter J. Artymiuk. 2014. COGNAC: a web server for searching and annotating hydrogen-bonded base interactions in RNA three-dimensional structures. Nucleic Acids Res. 42 (W1): W382-W388.
Figure above presents the tree graphs used by COGNAC to represent hydrogen bond connected base interactions where R denotes a base residue position (node) and the edges of the graphs are the lines connecting the nodes for: (A) triples (B) quadruples (C) quintuples (D) sextuples.
Reference
1) Mohd Firdaus-Raih, Hazrina Yusof Hamdani, Nurul Nadzirin, Effirul Ikhwan Ramlan, Peter Willett and Peter J. Artymiuk. 2014. COGNAC: a web server for searching and annotating hydrogen-bonded base interactions in RNA three-dimensional structures. Nucleic Acids Res. 42 (W1): W382-W388.
There are three methods for searching motifs in InterRNA: (1) Search by motif type, (2) Search by PDB ID, and
(3) Search by keyword.
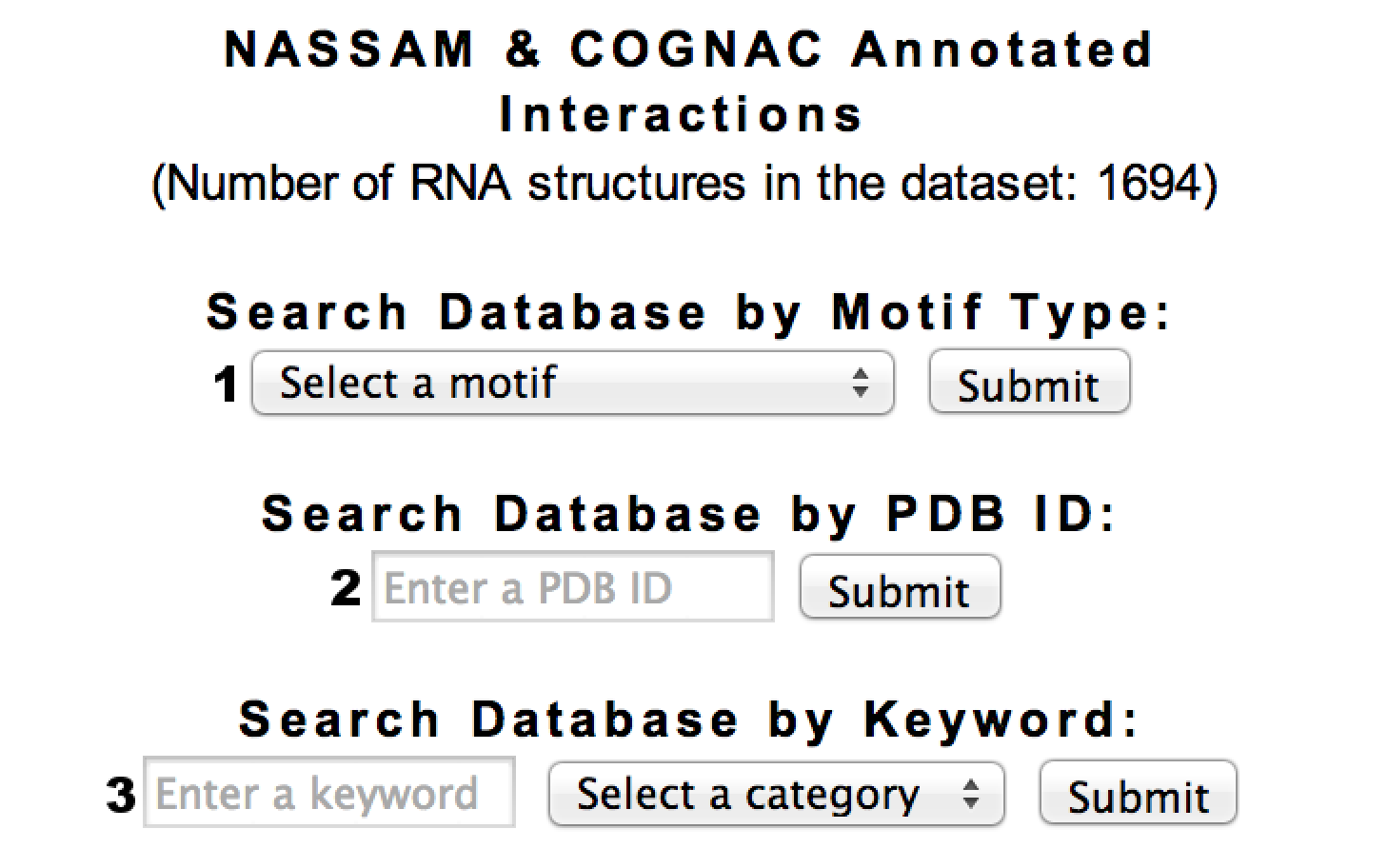
Each motif type is provided a page that lists the occurrences of that particular motif.
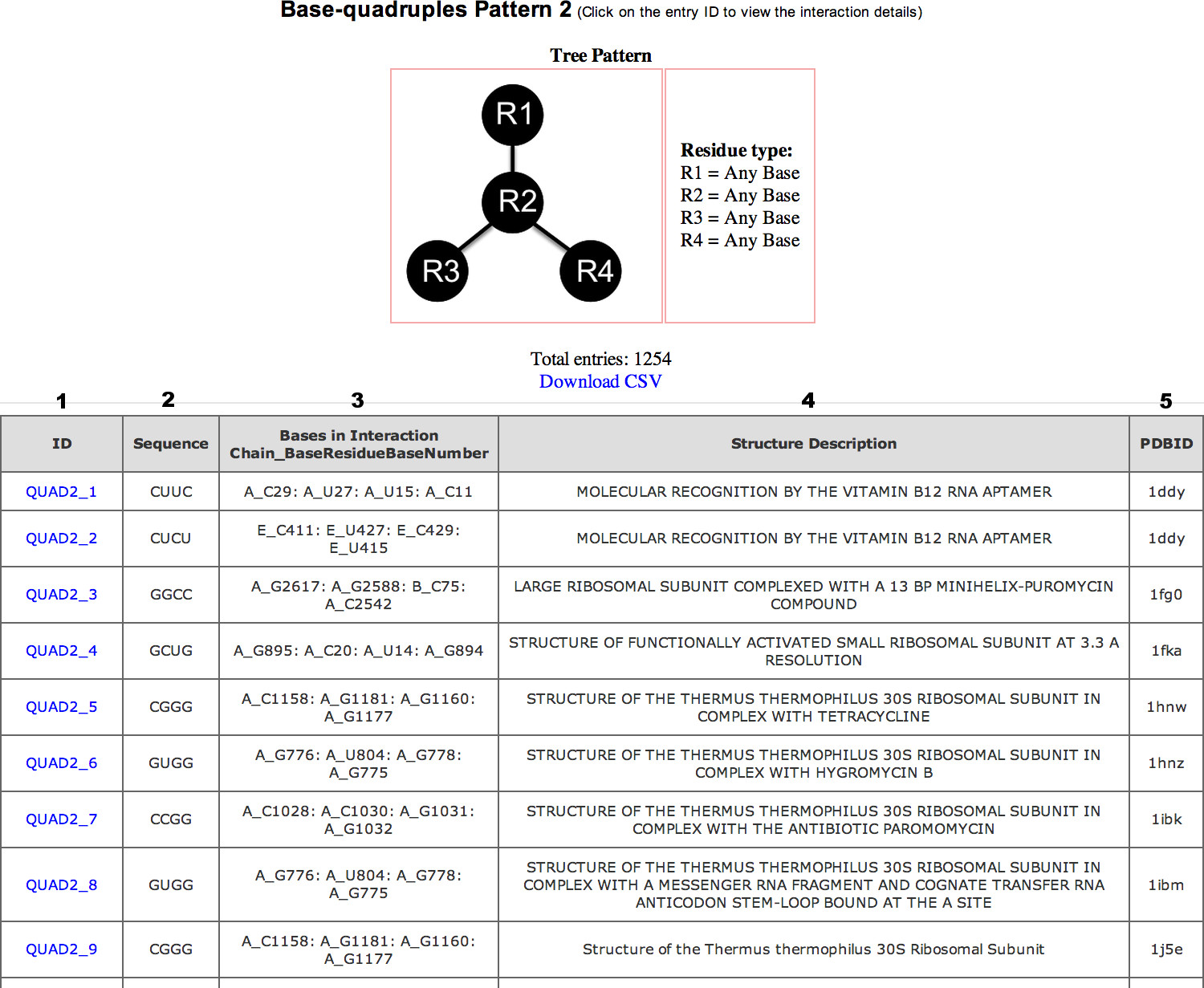
The motif type table consists of the following columns:
Users can select specific interaction by clicking on the entry ID. This will open up a new page that will provide (1) motif details, (2) list of hydrogen bond interactions in the motif and (3) Jsmol visualization options.
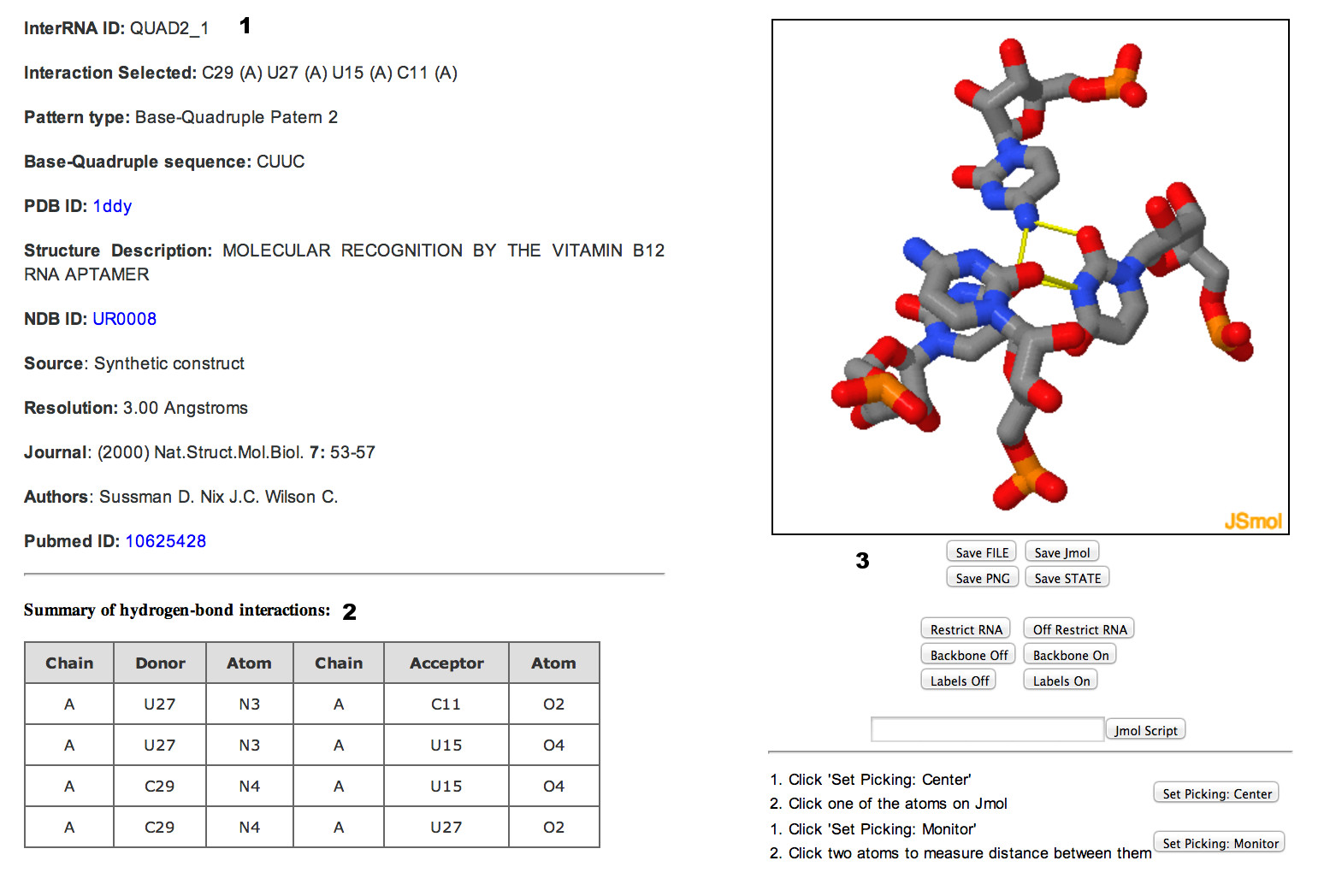
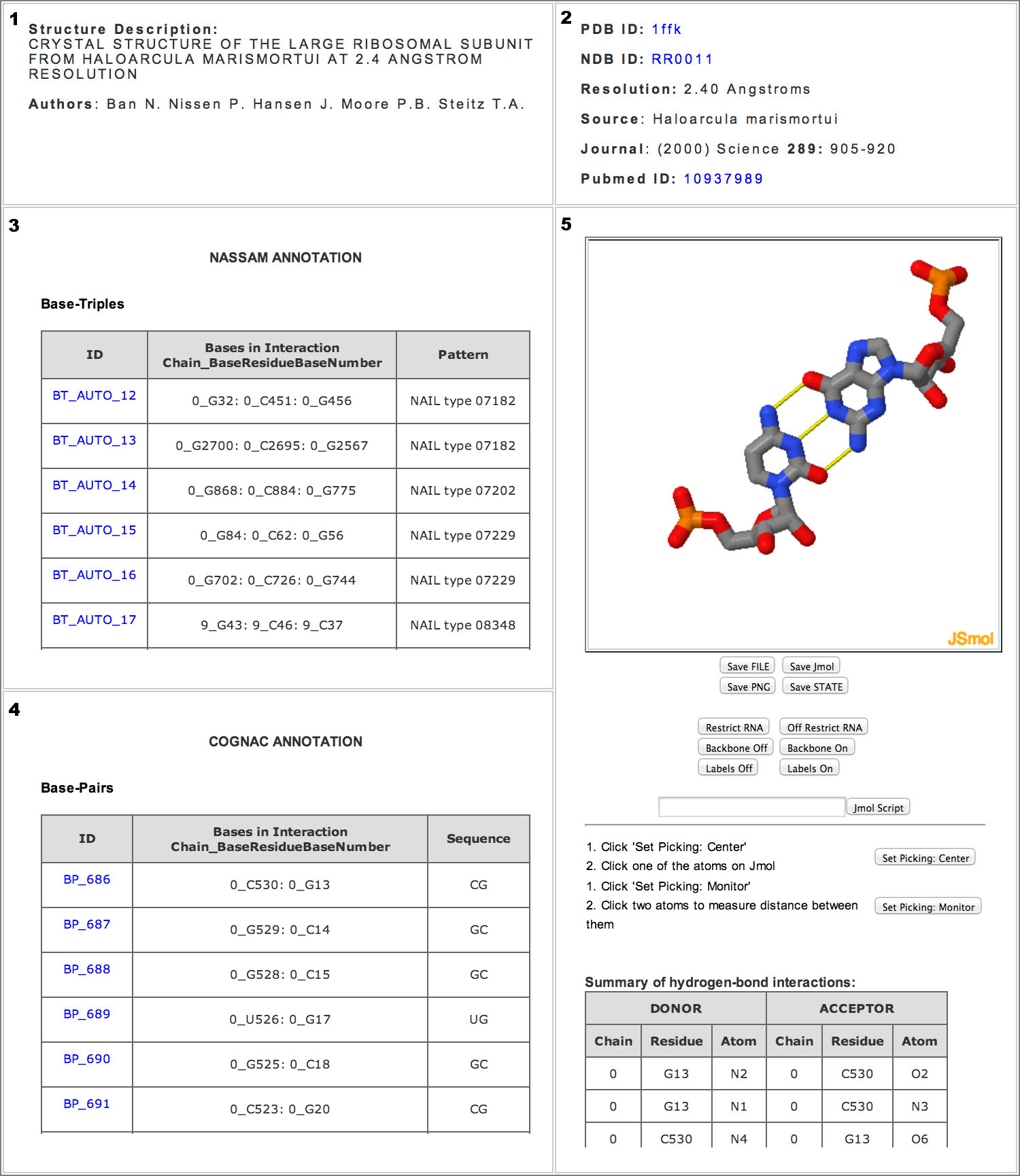
Users can also view all the motifs that are found in a particular RNA structure by entering the PDB ID of interest. This will open up a new page that will display (1 & 2) Structure details, (3) NASSAM annotations, (4) COGNAC annotations and (5) Jsmol visualization options. The hydrogen bond details will be presented under the Jsmol visualization window (5) when the user clicks on a specific NASSAM or COGNAC entry.
The search by keyword option is provided for five different categories: (1) PubMeD ID, (2) NDBID, (3) Author, (4) Resolution and (5) Organism. Please note of the following instructions when selecting the Resolution or Organism category.
Some operating systems have been known to block the use of Java due to security concerns and Jmol use may also be inconvenient to some users due to the requirement of needing Java installed for the browser. Therefore, InterRNA uses the Javascript implementation of Jmol (Jsmol) for RNA 3D motifs viewing. The Jsmol applet provide the following visualization options:
The download data option is provided for each motif type data. Currently, users can download the data in csv format. The order of columns in the csv file is as follows:
Please note that the number of columns for both Residue_Number and Residue_Chain varies according to the size of the motif. For example, in a base-triple interaction, there will be 3 columns for both Residue_Number and Residue_Chain whereas for a base-sextuple intereaction, there will be 6.









