To provide examples on how the web server can be used for protein binding site prediction, we illustrate some case studies used to analyse protein-drug interactions in different perspectives.
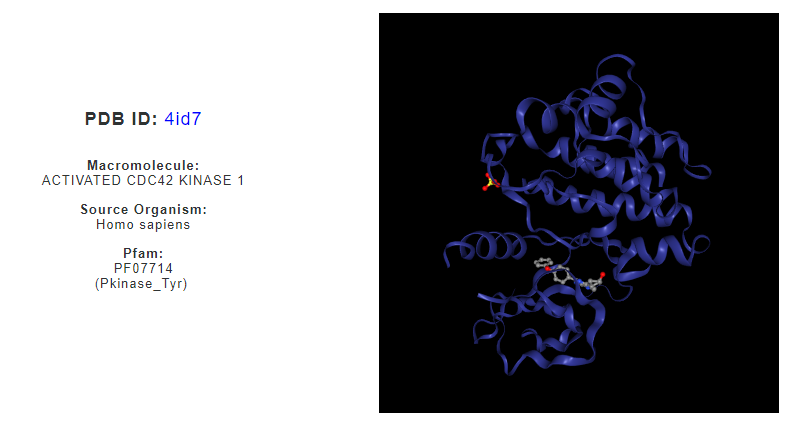
Activated Cdc42-associated kinase (ACK1) protein is a type of non-receptor tyrosine kinase, and is known for its binding to GTPase Cdc42. The protein involves in signalling and phosphorylation process and had recently been investigated for potential drug target related to cancer, due to its potential to remove tumor suppressor from cancer cells using similar mechanism. One of PDB structure of ACK1 is from Homo sapeins (PBD ID: 4id7) that is bound to cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol (PDB Ligand ID: 1G0), an ACK1 inhibitor. It is of interest to know other drug molecules that could bind to this protein. The web server can be used to search for possible drug molecule that can bind to ACK1 protein through structural similarity searches.
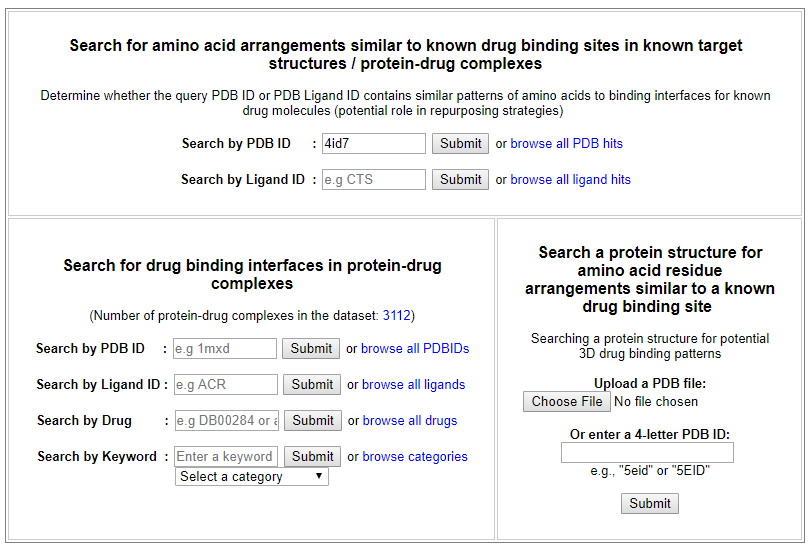
From the ‘Search for amino acid arrangements similar to known drug binding sites in known target structures / protein-drug complexes’ interface, PDB ID search for ‘4id7’ returns structural details and visualization of the protein structure, as well as a list of similar patterns of amino acids derived from ASSAM searches. User may filter the list based on RMSD, Z-score and sequence identity values, or select only matches from human structures only.
(Link)
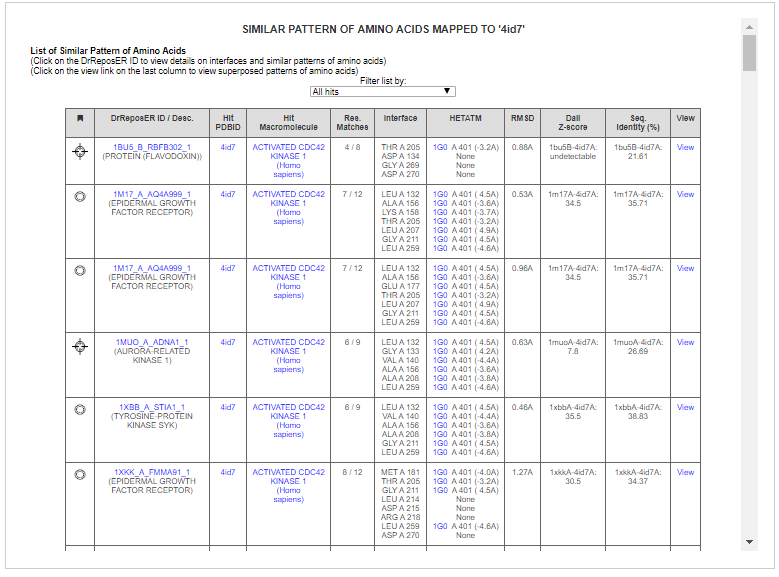
User can click on the DrReposER ID to get more details related to known drug binding sites having similar patterns of amino acids as the query structure.
Clicking the 'View' link on the last column will shows superposed patterns of amino acids in the NGL viewer.
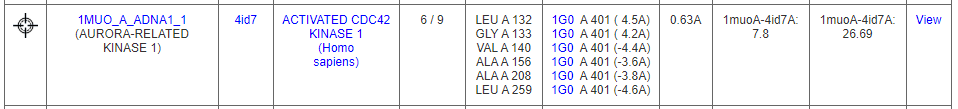
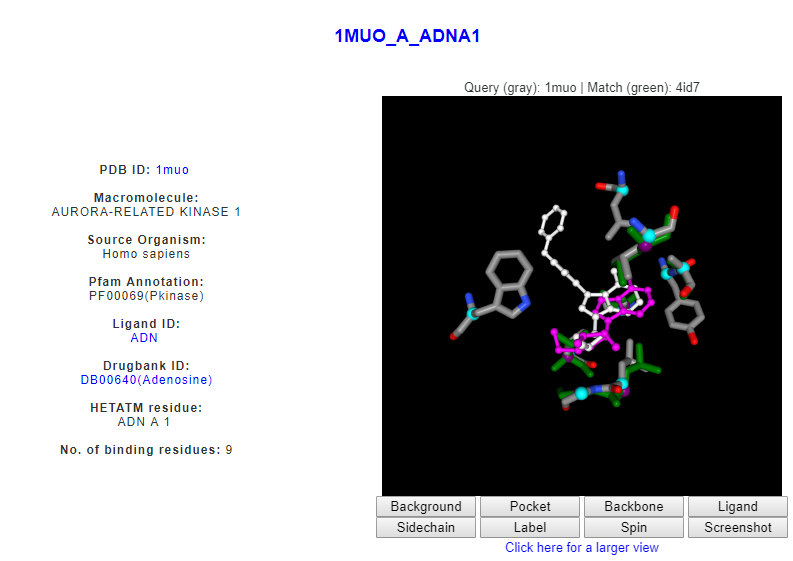
For example, the query protein, ACK1 is found to have similar pattern of amino acids with an Aurora-related protein kinase (DrReposER ID:1MUO_A_ADNA1) that is bound to Adenosine (PDB Ligand ID: ADN). Adenosine is a small molecule that acts as a neurotransmitter and involves in many biological roles.
(Link)
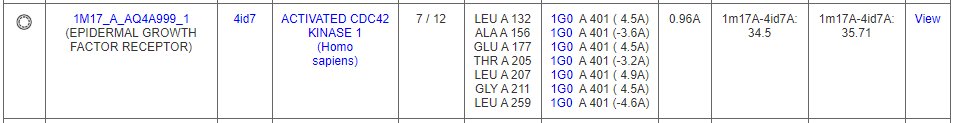
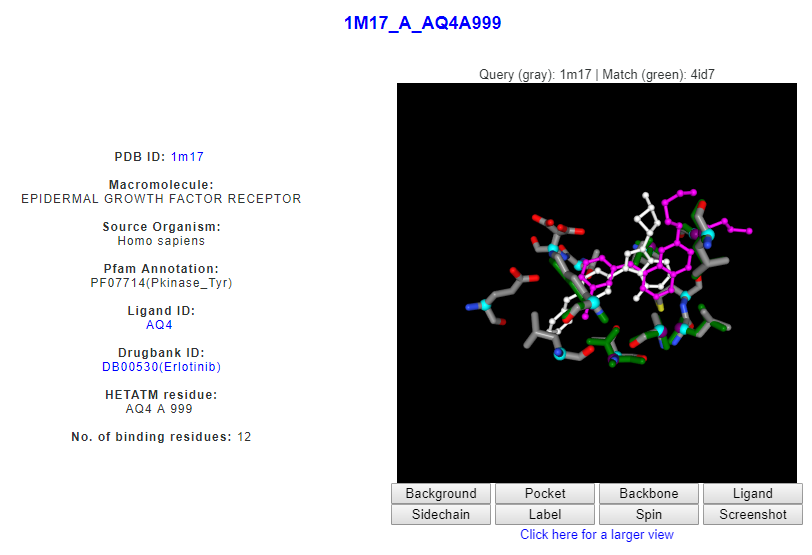
Another example is a similar pattern of amino acids with an epidermal growth factor receptor (DrReposER ID:1M17_A_AQ4A999) that is bound to Erlotinib (PDB Ligand ID: AQ4). Erlotinib has been indicated for non-small cell lung cancer, pancreatic cancer and several other type of cancer, thus can be proposed for potential binding to ACK1 protein.
(Link)
Other drug molecules that can potentially bind to ACK1 protein based on structural similarity of their binding sites include;
Lapatinib (FMM)-indicated for breast and lung cancer; Imatinib (STI)-indicated for various cancers including leukaemia and gastrointestinal stromal tumor; Dasatinib (1N1) and Nicotinib (NIL)-indicated for chronic myelogenous leukaemia.
In similar manner, searching for similar patterns of amino acids can be performed from PDB Ligand ID search. For example, searching for '1G0' on the interface will returns list of matched protein structures that is bound to IGO.
(Link)
References:
1) Jin, M. et al. (2013). Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors. Bioorganic & Medicinal Chemistry Letters. 23(4):979-984. (link)
1) Mahajan, K. & Mahajan, N. P. (2013). ACK1 Tyrosine Kinase: Targeted Inhibition to Block Cancer Cell Proliferation. Cancer Lett. 338(2):185-192. (link)







